MedMap
“MedMap,” is an Artificial Intelligence (AI) tool able to track with a high degree of semantic precision unique medical cases, and analyze multiple cases with unambiguous, standards-based labels. This may include the early cases of a novel pathogen, as well as the finely differentiated paths in disease progression and stratification.
An ontology-based tagging, tag-suggestion and diagramming tool, MedMap leverages medical and everyday life ontologies, and the data they contain, suggesting labels with a high degree of semantic precision for supervised machine learning, and offering conjectures about possible labels in the case of unsupervised machine learning. In computer science, this project supplements the algorithmic power of machine and deep learning with under-exploited areas of data modelling, ontologies and semantics. Applied to the domain of medical informatics, this aligns with today’s objectives in the areas of precision or personalized medicine.
The semantic processes we have developed will enrich data and AI across populations at any scale by making datapoints smaller, more precise, and their relations specified with less ambiguity. Cross-mapping between ontologies by users will progressively yield further precision. The accumulated knowledge of single cases will generate a richer set of annotated training data to further enable the use of powerful supervised learning approaches to further extract knowledge and help address similar scenarios in the future.
The figure below illustrates a working prototype created to demonstrate how MedMap works. On the left is medical data, which may be one of the three possible sites of application we discuss below: 1) a deeply documented, peer reviewed single clinical case; 2) a smart medical record; or 3) a raw dataset of any size extracted from a medical information system as structured or unstructured text.
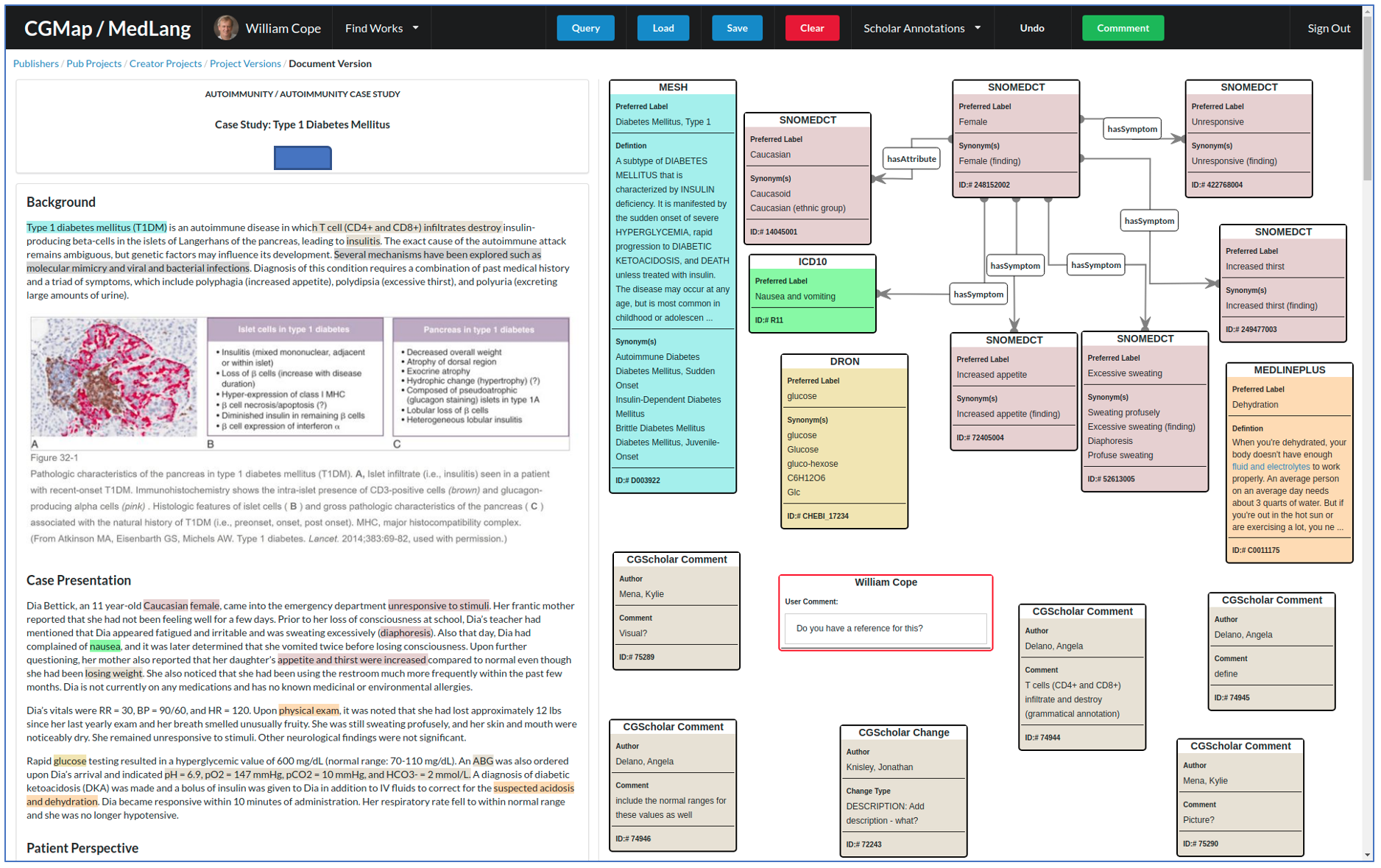
Left side: multimodal case documentation with color-coded annotations according to ontology items in the visualization. Right side: A medical logic model visualization, where each node and relation is linked to a standard medical ontology, e.g. International Statistical Classification of Diseases (ICD); the International Classification of Functioning, Disability and Health (ICF); the Systematized Nomenclature of Medicine (SNOMED); Logical Observation Identifiers Names and Codes (LOINC); and The Drug Ontology (DRON), as well as everyday life ontologies operating at a high level of semantic precision, e.g. place (GeoNames), time and event (iCal), demographic profile (age, gender, race/ethnicity etc.), occupational classification (SIC: Standard Occupational Classification), or objects in the form of identifiable products (IAN: International Article Number) – and many others.
This project responds builds on several longstanding lines work, including e-learning ecologies and the theory and practice of ontology and metaontology in our transpositional grammar. This work has been supported by US Department of Education, Gates Foundation and NSF grants. MedMap has been developed with support of a Jump ARCHES grant from the Healthcare Engineering Research Center at the University of Illinois.
Applications
Among a number of possibilities, we anticipate the following potentially applications:
- Medical Education. Medical and health sciences students create clinical cases using MedMap as a suggestion and concept mapping tool (the illustration above).
- Single clinical case documentation. This will fill a gap in medical science research and publication, which currently tends to prioritize larger population sizes. Semantically rich, single case documentation will highlight puzzling and worrisome cases, or cases of a kind frequently encountered but requiring more detailed documentation to differentiate variable disease causes and progressions. Using highly granular medical and everyday ontologies, multiple cases could be datamined for outlier warnings, as well as shared features.
- A smart medical record. This will be a shadow box overlay or parallel screen where: a) the content of a medical record can be more precisely specified by node selection from medical and everyday ontologies, then logic models created to assist the clinician in their thinking. The amount of extra work for the medical professional would be minimized. Effort would more than rewarded by smart node suggestions made by the system and the clinical logic and decision modeling by diagramming nodes in the visualization.
- Adding semantic precision to medical datasets. This application of the MedMap tool would support searching for already labelled datasets based on the salience of semantic connections; requesting or suggesting labels based on outlier alerts, or identifying semantically germane patterns in smaller or larger populations; and generating more semantically accurate topic models.
Paper
- Cope, Bill, Mary Kalantzis, ChengXiang Zhai, Andrea Krussel, Duane Searsmith, Duncan Ferguson, Richard Tapping and Yerko Berrocal. 2022. "Maps of Medical Reason: Applying Knowledge Graphs and Artificial Intelligence in Medical Education and Practice." in Bioinformational Philosophy and Postdigital Knowledge Ecologies, edited by M. Peters, P. Jandrić and S. Hayes. Cham CH: Springer.
